The Ovarian Pre-cancer Atlas is developed by the Shih Laboratory at Johns Hopkins University to share spatial transcriptomic insights on precursor lesions of ovarian cancer. Our goal is to provide the research community with intuitive tools to explore gene expression patterns across the fallopian tube epithelium and associated precancerous lesions.
You may search genes of interest to inquire mRNA expression levels and compare them in normal fallopian tube epithelium, high-grade serous carcinoma, and various types of precancerous lesions including STIC and p53 signatures. In the forthcoming version, the integrated morphology, multi-omic data, and relevant clinical information will be available.
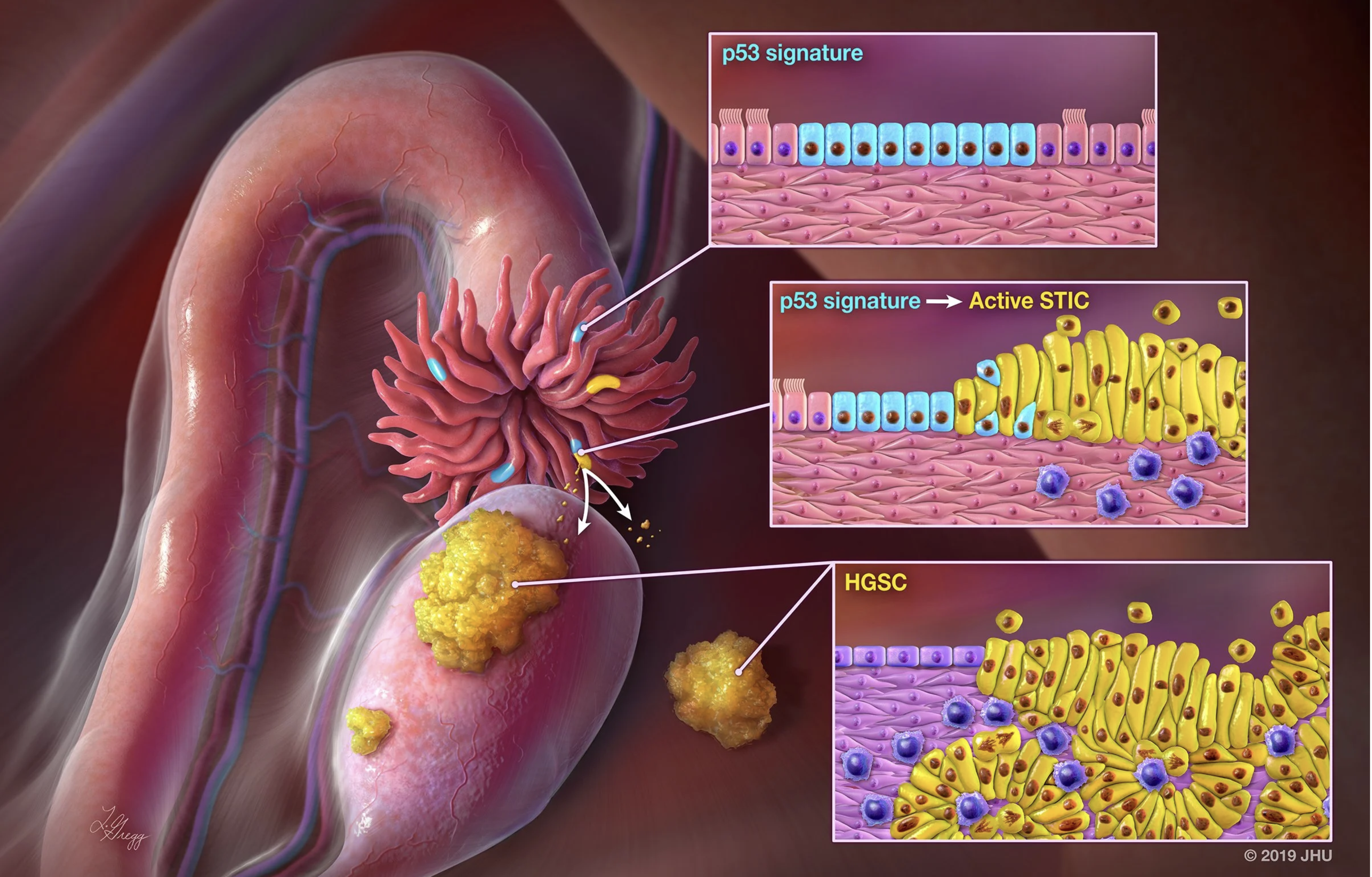
Gene Expression across Ovarian Precancerous Lesions
Explore modeled RNA expression across normal fallopian tube epithelium, precursor lesions, and high-grade serous carcinoma using NanoString GeoMx DSP data.
💡 How to Use
- Select Type: Choose Epithelium or Stroma to define the tissue compartment.
- Enter Gene: Type an official gene symbol (e.g., SOX4, MMP7, CD74).
- (Optional) Select Category: Add an additional plot to compare STIC separated by morphology category (Flat and BLAD).
- Click Generate Plot: Boxplots will display expression levels across lesion groups.
Note on “*”: The asterisk marks groups whose group mean is above or below the NFT (normal fallopian tube epithelium) mean based on a simple reference comparison computed in Python. It is a visual cue only and does not indicate statistical significance (no p-value). For formal differential expression or significance testing, please use DESeq2 (or an equivalent method).
Gene Correlation across Ovarian Precancerous Lesions
Explore the relationship between two genes across selected lesion groups using mRNA expression data from NanoString GeoMx spatial transcriptomics.
💡 How to Use
- Select Category: Choose a grouping variable (e.g., Diagnosis, Morphology category, or Molecular subtype).
- Select Group(s): Choose one or multiple lesion groups to include in the correlation analysis.
- Color By (Optional): Highlight points by Diagnosis or Molecular Subtype.
- Enter Gene A and Gene B: Input two official gene symbols (e.g., IGFBP2 and MMP7).
- Click “Generate Correlation”: A scatter plot will display gene expression correlation across the selected groups.
